Achievement
In this paper, we propose a novel privacy-preserving Deep Learning model distribution across cancer registries for information extraction from cancer pathology reports with privacy and confidentiality considerations. The proposed approach exploits the adversarial training framework to distinguish private features from shared features among different datasets. It only shares registry-invariant model parameters, without sharing raw data nor registry-specific model parameters among cancer registries. Thus, it protects both the data and the trained model simultaneously. We compare our proposed approach to single-registry models, and a model trained on centrally hosted data from different cancer registries. The results show that the proposed approach significantly outperforms the single-registry models and achieves statistically indistinguishable micro and macro F1-scoreas compared to the centralized mode.
Significance and Impact
Collaboration among cancer registries is essential to develop accurate, robust, and generalizable deep learning models for automated information extraction from cancer pathology re-ports. Sharing data presents a serious privacy issue, especially in biomedical research and healthcare delivery domains. Distributing pretrained deep learning (DL) models has been proposed to avoid critical data sharing. However, there is growing recognition that collaboration among clinical institutes through DL model distribution exposes new security and privacy vulnerabilities. These vulnerabilities increase in natural language processing (NLP) applications, in which the dataset vocabulary with word vector representations needs to be associated with the other model parameters.
Research Details
- The datasets used in this paper are 374,899 and 172,128 pathology reports obtained from two independent SEER program sources: the Louisiana Tumor Registry (LTR) and Kentucky Cancer Registry (KCR) respectively. The LTR corpus spans the period 2004-2018 while the KCR corpus spans the period 2009-2018.
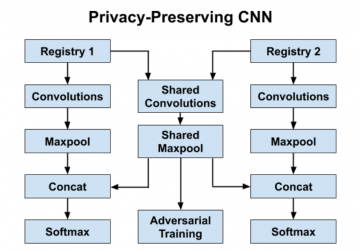
- We use three parallel 1-D convolution layers which respectively examine three, four, and five consecutive words at a time – these layers act as feature extractors which identify important combinations of words for a given task. We concatenate the outputs of these convolutions and then use a temporal maxpool to filter the most salient word combinations generated by each convolution filter. The final selected features represent the most important word n-grams in the document for the task at hand; this is fed into a softmax layer for classification.
- Our privacy-preserving CNN utilizes registry-specific layers for each registry and shared layers for all registries; our goal is to ensure that the registry-specific layers learn features private to that specific registry dataset, while the shared layers learn registry-agnostic features from all registry datasets.
- To show the performance of data sharing and model distribution approaches on tackling the class imbalance problem, which is common in clinical datasets, we compute the F1- score per class label of all models. Since we have hundreds of subsite labels, we illustrate the performance of different training approaches on the most and least represented classes with at least 100 samples. The results clearly show that non-private data sharing, the centralized model, and privacy-preserving model distribution approaches improve the classification accuracy of low prevalent class labels. However, all models perform equally well on the more prevalent classes
Overview
In this paper, we propose a privacy-preserving model distribution across cancer registries technique. It shares the registry-agnostic DL model parameters across cancer registries, and excludes any registry-specific parameters that may compromise privacy violations. The proposed method eliminates the need of a centralized host for datasets and the need of complex encryption techniques to privately distribute DL models across the collaborating institutes. The experiments demonstrate that our proposed DL training approach significantly out performs the single-registry model and achieves a comparable performance to the centralized model. Future directions of this work include applying the proposed approach on more cancer registries and information extraction tasks as well as using the proposed approach to securely distribute deep learning model across registries through recently developed model distribution techniques, such as cyclical weight transfer.
Last Updated: May 28, 2020 - 4:01 pm